


Go long.
Get the full picture of the genome with DeCodi-Fi™
DeCodi-Fi™ is a high-fidelity DNA polymerase designed to amplify long DNA fragments, up to 44 kb, with exceptional fidelity, high yield, fast cycling, and robust performance from ultra-low input.
Optimized for next-generation applications such as long-read sequencing, whole-genome assembly, and structural variant detection, it delivers the length and accuracy required even when DNA is limited, bridging the gap between low input and long-range success.

Go long.
Get the full picture of the genome with DeCodi-Fi™
DeCodi-Fi™ is a high-fidelity DNA polymerase designed to amplify long DNA fragments, up to 44 kb, with exceptional fidelity, high yield, fast cycling, and robust performance from ultra-low input.
Optimized for next-generation applications such as long-read sequencing, whole-genome assembly, and structural variant detection, it delivers the length and accuracy required even when DNA is limited, bridging the gap between low input and long-range success.
-
Longer fragments: Amplifies up to 44 kb, delivering high-yield templates ready
for long-read sequencing platforms. -
Low input compatibility: Amplifies complex DNA from as little as 0.1 ng and medium-complexity templates from 5 pg.
-
High-Fidelity: More than 60% reduction in error rates, delivering high-accuracy amplification for precision-critical applications.
-
High yield, low bias: Uniform amplification and high DNA recovery, even from challenging or GC-rich templates. (25%-85% GC content)
-
Fast cycling: Achieves rapid amplification with extension speeds under 15 sec/kb.
- Uracil tolerance for epigenetic workflows: Efficiently reads through uracil, supporting both methylation analysis and protocols involving damaged DNA.
-
Amplification of DNA fragments for cloning
-
Amplification of long or GC rich templates
-
Library amplification for sequencing
-
High Fidelity and Specificity PCR
-
High-throughput PCR
Technical Data Sheets
![]() DeCodi-Fi All-In-One Mix TDS / Protocol
DeCodi-Fi All-In-One Mix TDS / Protocol
![]() DeCodi-Fi PCR Kit TDS / Protocol
DeCodi-Fi PCR Kit TDS / Protocol
Brochure
![]() DeCodi-Fi High-Fidelity Polymerase Brochure
DeCodi-Fi High-Fidelity Polymerase Brochure
Posters
![]() Unveiling Microbial Diversity Cost-Effective 16S rRNA Amplicon Sequencing using DeCodi-Fi High Fidelity Polymerase
Unveiling Microbial Diversity Cost-Effective 16S rRNA Amplicon Sequencing using DeCodi-Fi High Fidelity Polymerase
![]() DeCodi-Fi: A Cost-Effective Polymerase for High-Quality Genome Assembly of Marine Sediment Bacteria
DeCodi-Fi: A Cost-Effective Polymerase for High-Quality Genome Assembly of Marine Sediment Bacteria
![]() Solving amplification of high-GC expanded repeats to enable rare disease detection using long-read sequencing
Solving amplification of high-GC expanded repeats to enable rare disease detection using long-read sequencing
![]() A long-read 16S rRNA sequencing workflow enhances bacterial diversity recovery from rhizospheric soils of a salt flat in the Atacama desert
A long-read 16S rRNA sequencing workflow enhances bacterial diversity recovery from rhizospheric soils of a salt flat in the Atacama desert
Application Notes
![]() Resistance of High-Fidelity DNA polymerases to magnetic beads' presence
Resistance of High-Fidelity DNA polymerases to magnetic beads' presence
![]() Direct PCR from Crude Plant Extracts Using DeCodi-Fi High-Fidelity Polymerase
Direct PCR from Crude Plant Extracts Using DeCodi-Fi High-Fidelity Polymerase
![]() DeCodi-Fi in Amplicon Sequencing: A Sensitive High-Fidelity Polymerase for Xanthomonas citri Pathovar Differentiation
DeCodi-Fi in Amplicon Sequencing: A Sensitive High-Fidelity Polymerase for Xanthomonas citri Pathovar Differentiation
![]() Oxford Nanopore 16S Sequencing of Soil Samples from the Atacama Desert
Oxford Nanopore 16S Sequencing of Soil Samples from the Atacama Desert
![]() Comparative analysis of size bias in PCR Multiplex amplification using DeCodi-Fi™ vs commercial HiFi polymerases
Comparative analysis of size bias in PCR Multiplex amplification using DeCodi-Fi™ vs commercial HiFi polymerases
Quick Start Guides
![]() DeCodi-Fi All-In-One Mix Quick Start Guide
DeCodi-Fi All-In-One Mix Quick Start Guide
![]() DeCodi-Fi PCR Kit Quick Start Guide
DeCodi-Fi PCR Kit Quick Start Guide
Citations
![]() Clinical Chemistry - B-224 Long read sequencing: High-fidelity amplification of disease-associated complex genomic regions using DeCodi-Fi polymerase
Clinical Chemistry - B-224 Long read sequencing: High-fidelity amplification of disease-associated complex genomic regions using DeCodi-Fi polymerase
-
Longer fragments: Amplifies up to 44 kb, delivering high-yield templates ready
for long-read sequencing platforms. -
Low input compatibility: Amplifies complex DNA from as little as 0.1 ng and medium-complexity templates from 5 pg.
-
High-Fidelity: 64% higher fidelity than KAPA HiFi, reducing error rates in
high-precision applications. -
High yield, low bias: Uniform amplification and high DNA recovery, even from challenging or GC-rich templates. (25%-85% GC content)
-
Fast cycling: Achieves rapid amplification with extension speeds under 15 sec/kb.
- Uracil-friendly for epigenetic workflows: Reads through uracil, supporting methylation studies and damage-tolerant protocols.
-
Amplification of DNA fragments for cloning
-
Amplification of long or GC rich templates
-
Library amplification for sequencing
-
High Fidelity and Specificity PCR
-
High-throughput PCR
Technical Data Sheets
![]() DeCodi-Fi All-In-One Mix TDS / Protocol
DeCodi-Fi All-In-One Mix TDS / Protocol
![]() DeCodi-Fi PCR Kit TDS / Protocol
DeCodi-Fi PCR Kit TDS / Protocol
Brochure
![]() DeCodi-Fi High-Fidelity Polymerase Brochure
DeCodi-Fi High-Fidelity Polymerase Brochure
Posters
![]() Unveiling Microbial Diversity Cost-Effective 16S rRNA Amplicon Sequencing using DeCodi-Fi High Fidelity Polymerase
Unveiling Microbial Diversity Cost-Effective 16S rRNA Amplicon Sequencing using DeCodi-Fi High Fidelity Polymerase
![]() DeCodi-Fi: A Cost-Effective Polymerase for High-Quality Genome Assembly of Marine Sediment Bacteria
DeCodi-Fi: A Cost-Effective Polymerase for High-Quality Genome Assembly of Marine Sediment Bacteria
![]() Solving amplification of high-GC expanded repeats to enable rare disease detection using long-read sequencing
Solving amplification of high-GC expanded repeats to enable rare disease detection using long-read sequencing
![]() A long-read 16S rRNA sequencing workflow enhances bacterial diversity recovery from rhizospheric soils of a salt flat in the Atacama desert
A long-read 16S rRNA sequencing workflow enhances bacterial diversity recovery from rhizospheric soils of a salt flat in the Atacama desert
Application Notes
![]() Resistance of High-Fidelity DNA polymerases to magnetic beads' presence
Resistance of High-Fidelity DNA polymerases to magnetic beads' presence
![]() Direct PCR from Crude Plant Extracts Using DeCodi-Fi High-Fidelity Polymerase
Direct PCR from Crude Plant Extracts Using DeCodi-Fi High-Fidelity Polymerase
![]() DeCodi-Fi in Amplicon Sequencing: A Sensitive High-Fidelity Polymerase for Xanthomonas citri Pathovar Differentiation
DeCodi-Fi in Amplicon Sequencing: A Sensitive High-Fidelity Polymerase for Xanthomonas citri Pathovar Differentiation
![]() Oxford Nanopore 16S Sequencing of Soil Samples from the Atacama Desert
Oxford Nanopore 16S Sequencing of Soil Samples from the Atacama Desert
![]() Comparative analysis of size bias in PCR Multiplex amplification using DeCodi-Fi™ vs commercial HiFi polymerases
Comparative analysis of size bias in PCR Multiplex amplification using DeCodi-Fi™ vs commercial HiFi polymerases
Quick Start Guides
![]() DeCodi-Fi All-In-One Mix Quick Start Guide
DeCodi-Fi All-In-One Mix Quick Start Guide
![]() DeCodi-Fi PCR Kit Quick Start Guide
DeCodi-Fi PCR Kit Quick Start Guide
Citations
![]() Clinical Chemistry - B-224 Long read sequencing: High-fidelity amplification of disease-associated complex genomic regions using DeCodi-Fi polymerase
Clinical Chemistry - B-224 Long read sequencing: High-fidelity amplification of disease-associated complex genomic regions using DeCodi-Fi polymerase
THE MOST AFFORDABLE HIGH-FIDELITY IN THE MARKET
DeCodi-Fi™ offers a cost-effective option to high-fidelity polymerases, delivering substantial value while maintaining superior performance.
THE MOST AFFORDABLE HIGH-FIDELITY IN THE MARKET
DeCodi-Fi™ offers a cost-effective option to high-fidelity polymerases, delivering substantial value while maintaining superior performance.
LONG AMPLIFICATION & HIGH YIELD +
Maximize target yield with highly efficient amplification, even with fragment sizes up to 44 kb of length.
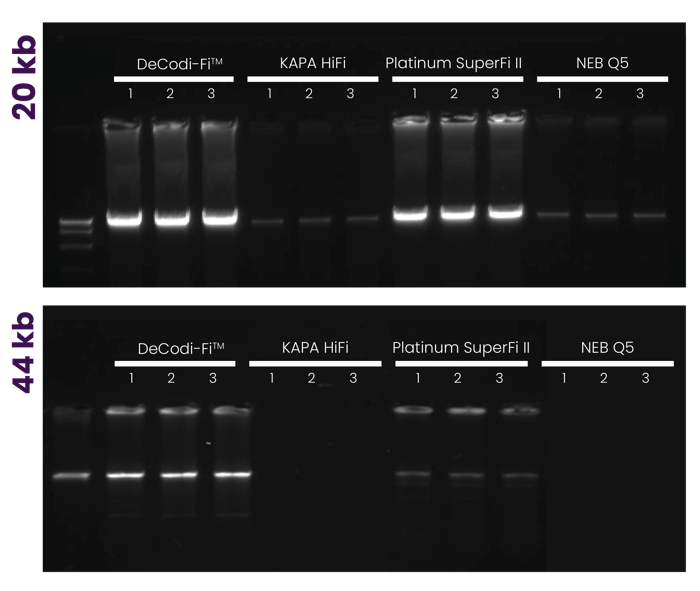
Amplification of lambda DNA fragments 20 kb and 44 kb with DeCodi-Fi™ High-Fidelity polymerase, KAPA HiFi, Platinum SuperFi II, and Q5. Each target was amplified from low input (1ng). Reactions were performed following the optimized conditions for each enzyme and using 24 cycles. The experiment was run in triplicate.
LOW INPUT +
Push the limits of long-read sequencing with low input. DeCodi-Fi™ amplifies long fragments from just 5 pg—where others fail even at 50 pg.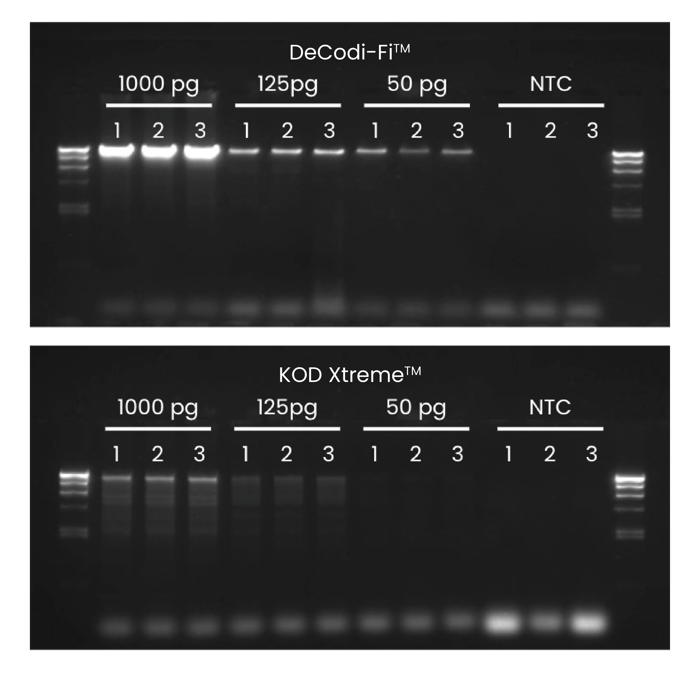
Amplification of the 17.6 kb human beta globin gene was performed using DeCodi-Fi™ High-Fidelity Polymerase and KOD Xtreme™ Hot Start DNA Polymerase. Each target was amplified from different inputs in a 25 μL reaction: 1000 pg, 125 pg, and 50 pg. A no-template control (0 pg) was also included. All reactions were performed following the optimized conditions for each enzyme and using 30 cycles. The experiment was run in triplicate.
HIGH-FIDELITY +
DeCodi-Fi High-Fidelity Polymerase exhibits a 64% reduction in error rates compared to KAPA HiFi.
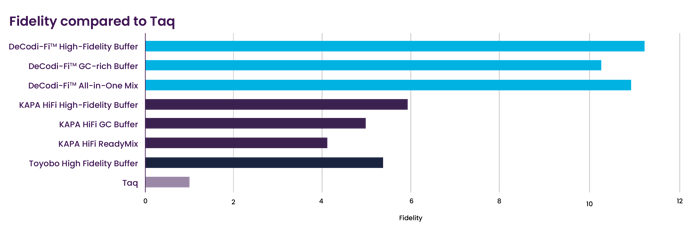
Comparison of DeCodi-Fi™, KAPA HiFi, and Toyobo HiFi, measured in terms of fold improvement over Taq Polymerase, based on Illumina libraries of the complete E. coli genome.
HIGH YIELD +
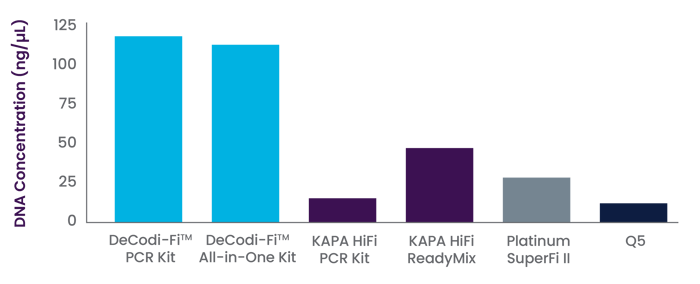
DNA concentration obtained from 25 μL of lambda 15 kb PCR reaction using DeCodi-Fi™ PCR kit, DeCodi-Fi™ All-in-One Mix, and competitors enzymes (Kapa HiFi, Platinum SuperFi II, and Q5). The experiment was run in triplicate.
SEQUENCE COVERAGE +
Ensures low bias amplification, even using different buffers and in AT-rich or GC-rich fragments, guaranteeing a more uniform coverage and improved sequencing depth.
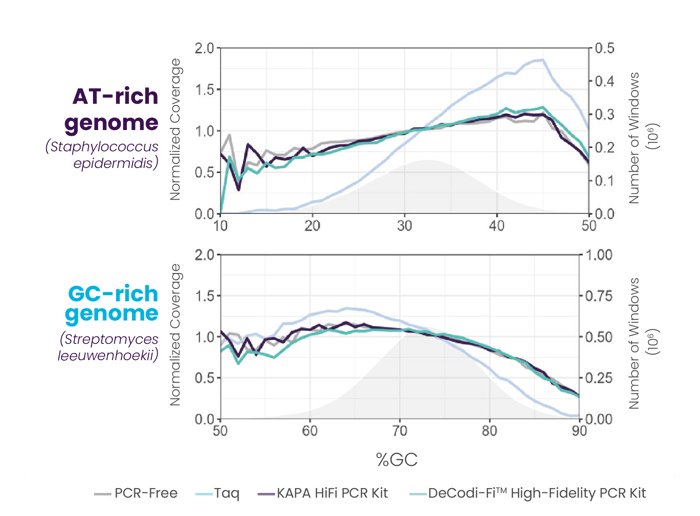
Illumina libraries prepared from a common genome (Escherichia coli) were amplified using Taq polymerase, KAPA HiFi PCR kit, and DeCodi-Fi™ High-Fidelity PCR kit (left). Illumina libraries prepared from an AT-rich genome (Staphylococcus epidermidis) and a GC-rich genome (Streptomyces leeuwenhoekii) were amplified using Taq polymerase, KAPA HiFi PCR kit, and DeCodi-Fi High-Fidelity PCR kit (right).
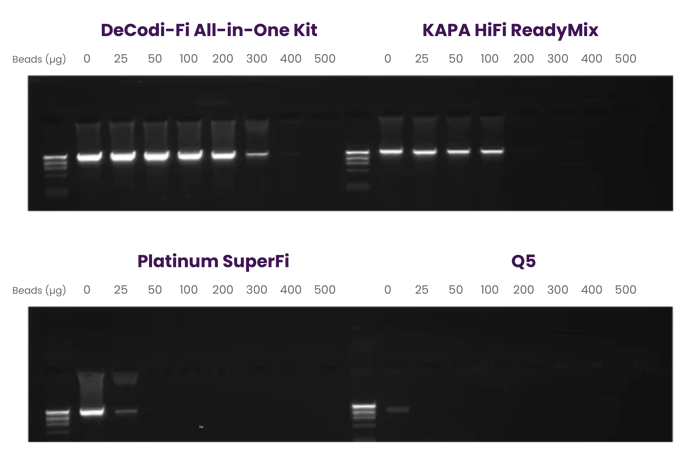
COMPATIBILITY WITH MAGNETIC BEADS +
DeCodi-Fi 2X shows the highest resistance to magnetic beads compared to competitors, excelling in long amplicon PCR. This makes DeCodi-Fi a consistent, robust, and affordable high-fidelity DNA polymerase for various methodologies requiring magnetic beads without compromising performance.
URACIL TOLERANCE +
DeCodi-Fi reads and robustly amplifies uracil-containing DNA.
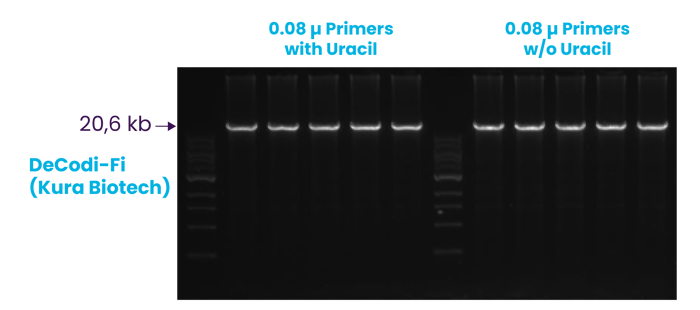
Robust amplification of a 20.6 kb λ DNA fragment from 2 ng of input DNA performed in pentaplicate, using primers with and without uracil, confirming comparable amplification efficiency and demonstrating tolerance to uracil-containing primers.
TWO FORMATS AVAILABLE
 DeCodi-Fi™
DeCodi-Fi™
2X All-In-One Mix
![]()
The All-in-One Mix format provides our hotstart high-fidelity polymerase in a user-friendly 2X MasterMix configuration. This format includes all the necessary components for the reaction (dNTPs, MgCl2, and stabilizers), excluding primers and template, in a proprietary buffer.
Product Type: All-in-One/MasterMix
Polymerase: DeCodi-Fi™ Hotstart High-Fidelity Polymerase
Format: 5 mL
Reactions: 400
Storage: Store at -20°C
 DeCodi-Fi™
DeCodi-Fi™
High-Fidelity PCR Kit
![]()
The High-Fidelity PCR Kit provides our hotstart high-fidelity polymerase and all the necessary components for the reaction separately for greater flexibility. It includes two buffers: a 5X High-Fidelity Buffer recommended for amplifying most templates which have balanced GC/AT content, and a 5X GC-rich Buffer recommended for amplifying GC-rich targets.
Product Type: PCR kit
Polymerase: DeCodi-Fi™ Hotstart High-Fidelity Polymerase
Reactions: 400
Storage: Store at -20°C
TWO FORMATS AVAILABLE
 DeCodi-Fi™
DeCodi-Fi™
2X All-In-One Mix
![]()
The All-in-One Mix format provides our hotstart high-fidelity polymerase in a user-friendly 2X MasterMix configuration. This format includes all the necessary components for the reaction (dNTPs, MgCl2, and stabilizers), excluding primers and template, in a proprietary buffer.
Product Type: All-in-One/MasterMix
Polymerase: DeCodi-Fi™ Hotstart High-Fidelity Polymerase
Format: 5 mL
Reactions: 400
Storage: Store at -20°C
 DeCodi-Fi™
DeCodi-Fi™
High-Fidelity PCR Kit
![]()
The High-Fidelity PCR Kit provides our hotstart high-fidelity polymerase and all the necessary components for the reaction separately for greater flexibility. It includes two buffers: a 5X High-Fidelity Buffer recommended for amplifying most templates which have balanced GC/AT content, and a 5X GC-rich Buffer recommended for amplifying GC-rich targets.
Product Type: PCR kit
Polymerase: DeCodi-Fi™ Hotstart High-Fidelity Polymerase
Reactions: 400
Storage: Store at -20°C
Need a Non Hot-Start Version?
DeCodi-Fi™ is also offered in a non hot-start version, made on request and customizable to your needs.
FAQs
DeCodi-Fi™
What is the extension time of DeCodi-Fi™? +
DeCodi-Fi™ typically requires 30 seconds to extend DNA fragments under 1 Kilobase (kb). For larger fragments, add 15 seconds(sec) per additional kilobase. For example, to amplify a 23 kb fragment should be 30 sec plus 15 sec x 22kb which is 360 seconds (6 minutes).
What is the input amount recommended for the reaction? +
DeCodi-Fi™ amplifies high-complexity DNA starting from 0.1 ng template and medium-complexity DNA starting from 5 pg.
Recommended input amounts are 5–50 ng for genomic DNA and less than 1 ng for templates smaller than 50 kb. To obtain amplification at 25 cycles, load more than 104 copies of the template, but do not exceed 100 ng per reaction (e.g., 35 ng of human genomic DNA corresponds to 104 copies).
What storage conditions are recommended for DeCodi-Fi™? +
Store the box at -20°C immediately upon receipt to maintain optimal kit performance and avoid repeated freeze-thaw cycles.
DeCodi-Fi™ is designed to be transported at temperatures between 2°C and 8°C without performance loss. Tests have confirmed the components remain stable under these conditions for at least 10 days during transport.
How does the HotStart formulation work? +
The polymerase is inactivated by an antibody until the initial cycle of thermal denaturation. This prevents non-specific priming during setup and initiation, reducing unwanted amplification products and improving overall reaction efficiency.
How many cycles are recommended for amplification? +
DeCodi-Fi™ polymerase has higher amplification efficiency than commonly used polymerases, even for longer amplicons. We recommend starting with 25 cycles for most PCR with inputs >104 copies of template. Always keep cycling under 30, particularly for amplicons >10kb. Shorter amplicons can tolerate overcycling better.
For amplifying Illumina sequencing libraries <600bp we recommend 12-14 cycles when starting from 1ng of template for reaching peak yield.
The number of cycles can range from 10 to 30, with 25 cycles as a good starting point. Optimization may be needed based on your specific template, primers, and application. Fewer cycles reduce the probability of errors and help minimize nonspecific products, smearing, and the accumulation of high molecular weight artifacts in agarose gels.
Can you provide custom solutions? +
Yes, we offer OEM and custom solutions that allow you to utilize our extensive expertise in enzyme optimization, buffer refinement and manufacturing.
What recommendation is important for the primer design? +
We recommend adding two phosphorothioate bonds at the 3' end of primers for avoiding 3' exonuclease degradation, given by the strong proofreading activity of DeCodi-Fi™. This is particularly important for shorter primers or when amplifying NGS libraries. 3' primer degradation could result in amplification of unspecific products. Oligonucleotide synthesis companies recognize an asterisk between the selected bases as the symbol for ordering a phosphorothioate bond.
Example P5 primer with 2 phosphorothioate bonds at the 3' end:
5'- AATGATACGGCGACCACC*G*A -3'
FAQs
DeCodi-Fi
What is the extension time of DeCodi-Fi™? +
DeCodi-Fi™ typically requires 30 seconds to extend DNA fragments under 1 Kilobase (kb). For larger fragments, add 15 seconds(sec) per additional kilobase. For example, to amplify a 23 kb fragment should be 30 sec plus 15 sec x 22kb which is 360 seconds (6 minutes).
What is the input amount recommended for the reaction? +
DeCodi-Fi™ amplifies high-complexity DNA starting from 0.1 ng template and medium-complexity DNA starting from 5 pg.
Recommended input amounts are 5–50 ng for genomic DNA and less than 1 ng for templates smaller than 50 kb. To obtain amplification at 25 cycles, load more than 104 copies of the template, but do not exceed 100 ng per reaction (e.g. 35 ng of human genomic DNA corresponds to 104 copies).
What storage conditions are recommended for DeCodi-Fi™? +
Store the box at -20°C immediately upon receipt to maintain optimal kit performance and avoid repeated freeze-thaw cycles.
DeCodi-Fi™ is designed to be transported at temperatures between 2°C and 8°C without performance loss. Tests have confirmed the components remain stable under these conditions for at least 10 days during transport.
How does the HotStart formulation work? +
The polymerase is inactivated by an antibody until the initial cycle of thermal denaturation. This prevents non-specific priming during setup and initiation, reducing unwanted amplification products and improving overall reaction efficiency.
How many cycles are recommended for amplification? +
DeCodi-Fi™ polymerase has higher amplification efficiency than commonly used polymerases, even for longer amplicons. We recommend starting with 25 cycles for most PCR with inputs >104 copies of template. Always keep cycling under 30, particularly for amplicons >10kb. Shorter amplicons can tolerate overcycling better.
For amplifying Illumina sequencing libraries <600bp we recommend 12-14 cycles when starting from 1ng of template for reaching peak yield.
The number of cycles can range from 10 to 30, with 25 cycles as a good starting point. Optimization may be needed based on your specific template, primers, and application. Fewer cycles reduce the probability of errors and help minimize nonspecific products, smearing, and the accumulation of high molecular weight artifacts in agarose gels.
Can you provide custom solutions? +
Yes, we offer OEM and custom solutions that allow you to utilize our extensive expertise in enzyme optimization, buffer refinement and manufacturing.
What recommendation is important for the primer design? +
We recommend adding two phosphorothioate bonds at the 3' end of primers for avoiding 3' exonuclease degradation, given by the strong proofreading activity of DeCodi-Fi™. This is particularly important for shorter primers or when amplifying NGS libraries. 3' primer degradation could result in amplification of unspecific products. Oligonucleotide synthesis companies recognize an asterisk between the selected bases as the symbol for ordering a phosphorothioate bond.
Example P5 primer with 2 phosphorothioate bonds at the 3' end:
5'- AATGATACGGCGACCACC*G*A -3'
Ready to amplify your research?
Take your DNA sequencing to the next level with DeCodi-Fi™. Contact our team for sales and technical support.
Ready to Amplify Your Research?
Take your DNA sequencing to the next level with DeCodi-Fi.
Contact our team for sales and technical support.
